
Phenotyping field scanner system
The Lemnatec Field Scanalyzer at the University of Arizona Maricopa Agricultural Center and USDA Arid Land Research Station in Maricopa, Arizona is the largest field crop analytics robot in the world. This high-throughput phenotyping field-scanning robot has a 30-ton steel gantry that autonomously moves along two 200-meter steel rails while continuously imaging the crops growing below it with a diverse array of cameras and sensors.

Automated controlled-environment phenotyping
The Bellwether Foundation Phenotyping Facility is a climate controlled 70 m2 growth house with a conveyor belt system for moving plants to and from fluorescence, color, and nearinfrared imaging cabinets. This automated, high-throughput platform allows repeated non-destructive time-series image capture and multi-parametric analysis of 1,140 plants in a single experiment.

Phenotyping sensors on Unmanned Aerial Vehicles
UAVs will repeatedly fly precision passes over crops to provide autonomous image surveys, with onboard sensors georegistering the data and stitching together the captured images based on vehicle positions. The use of UAVs will provide rapid coverage of areas that are much larger than what can be covered with ground-based systems.

Phenotyping sensors on piloted ground vehicles
Multiple vehicles outfitted with instruments that measure canopy height, temperature, and spectral reflectance at three wavelengths will be used to collect plot-level data georeferenced at centimeter accuracy. Plot scale data is relevant for traits that are strongly influenced by interplant competition and crop geometry such as light interception and canopy temperature.

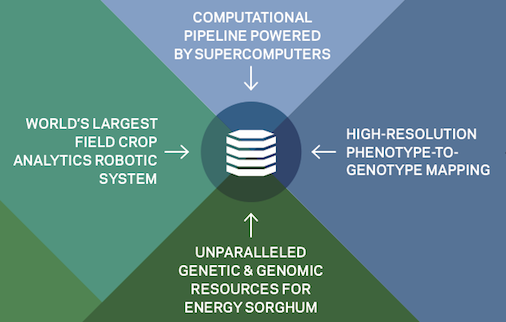
Genomic and genetic data and computational platform
We will conduct whole-genome resequencing on a diverse set of 400 sorghum accessions included in the Bioenergy Association Panel to measure the landscape of genetic variation in the selected germplasm and provide whole-genome sequences for association mapping. In addition, we will conduct genotyping by-sequencing on 400 lines from two recombinant inbred line (RIL) populations. The genomic data will then be used to identify the differences between each line and the reference genome sequence for sorghum. We will use bioinformatics and quantitative genetics to characterize the observed genetic variation and identify genomic regions controlling biomass, plant architecture, and photosynthetic traits.

High performance data storage and computing system
The system will provide public access to raw data, metadata, derived data, provenance for derived data, and standardized data processing workflows. This open-source compute platform will be used to provide open access to huge datasets that will guide breeding decisions, facilitate collaboration, and allow unprecedented data sharing.

 Field phenotyping field scanner system
Field phenotyping field scanner system Automated controlled-environment phenotyping
Automated controlled-environment phenotyping Phenotyping sensors on unmanned aerial vehicles
Phenotyping sensors on unmanned aerial vehicles Phenotyping sensors on piloted ground vehicles
Phenotyping sensors on piloted ground vehicles Genomic and genetic data and computational platform
Genomic and genetic data and computational platform High performance data storage and computing system
High performance data storage and computing system